The immune antibody library is produced either by the infected host or by immunity to the infectious agent. Antibodies from the immune library differ from those from the naive library because the host's internal immune mechanisms form the antibody library to produce high-affinity antibodies. Since the immune system evolves based on the health of the individual, the immune library can provide not only infection-specific antibodies but also antibodies from memory B cells, just like a naive library. Creative Biolabs has extensive research experience in several areas related to immune antibody libraries, including human antibody production by phage display immune library and single domain antibody library. Our aim is to provide you with a perfect scientific research service experience through professional service.
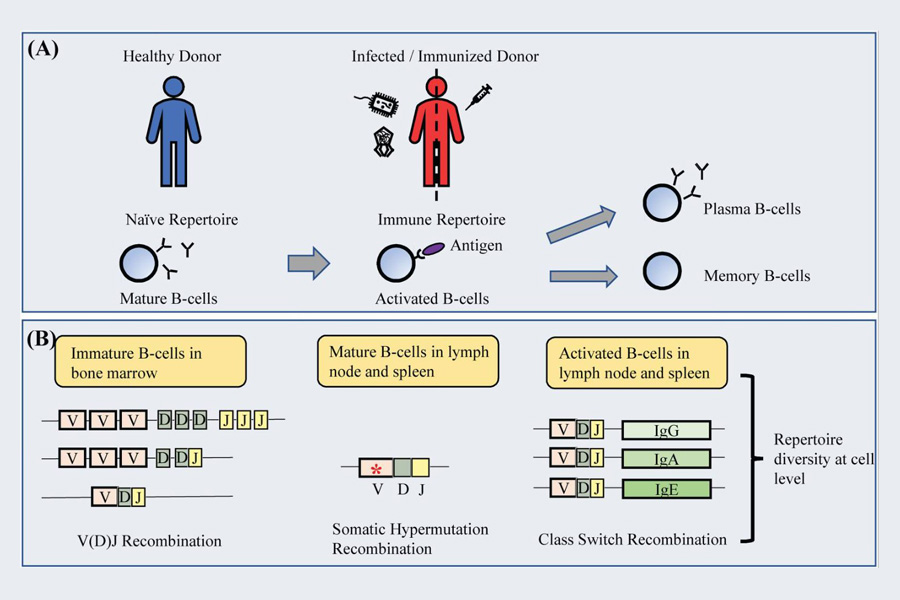 Fig 1. A. The antibody from mature B-cells of a healthy donor is referred to as a naïve repertoire. B. The natural antibody repertoire in a healthy individual is diversified via V(D)J recombination, and this represents a naïve antibody library repertoire. (Lai, J. Y., et al., 2020)
Fig 1. A. The antibody from mature B-cells of a healthy donor is referred to as a naïve repertoire. B. The natural antibody repertoire in a healthy individual is diversified via V(D)J recombination, and this represents a naïve antibody library repertoire. (Lai, J. Y., et al., 2020)
B cells are a major component of the immune system and protect the body from harmful antigens. B cells constantly function to produce antibodies in response to infections before and after they occur. The pool of antibodies in a healthy state is diverse enough to fight new infections while remembering old ones. This function of B cells is possible with the diverse repertoire attained from two primary mechanisms: V(D)J recombination of the variable (V), diversity (D), and joining (J) gene segments and somatic hypermutation (SHM). This includes, to a lesser extent, non-traditional secondary mechanisms that also increase the diversity of antibody libraries, including non-standard recombination in violation of the 12/23 recombination rule, SHM-associated genetic insertions and deletions, direct antigen contact in the non-complementary determining region (non-CDRs) of the antibody, post-translational modifications, conformational heterogeneity, and the use of non-protein cofactors. Together, these mechanisms contribute to various variations within antibody CDRs that form major antigen-binding sites. The events leading to the generation of mature antibody genes are also multifaceted, as the recombination of multiple variable genes provides a great deal of combinatorial diversity and is combined with different heavy (VH) and light (VL) chains. Since SHM and related mechanisms are triggered upon the encounter of antigens, exposure to infection affects the resulting library, as antibody libraries are biased and formed to fight the invading pathogen.
The main challenge in library construction is designing an effective strategy for antibody generation. Typically, libraries are generated using primers that target variable gene segments and link gene segments. However, without the use of isotype-specific reverse primers, it is difficult to differentiate between isotype responses.
The homotype-specific immune libraries have many obvious advantages. As B cells are activated, class-switching recombination (CSR) occurs, allowing antibody production in immature B cells to switch from IgM or IgD to isotype IgG, IgA, or IgE. IgG is further divided into one subclass (IgG2, IgG3, IgG4, and IgG1), while IgA is further divided into two subclasses (IgA30 and IgA31). The CSR event depends on the T helper cell response, which in turn depends on the nature of the antigen and the main invasion pathway. Each homotype has its own role and distribution sites in vivo, enabling effective combat against invasive antigens. For example, IgG is the most abundant contype in plasma and is responsible for preventing extracellular infections, while IgA is primarily found in mucous secretions and may reflect primary sensation on the mucosal surface. IgE is the immunoglobulin with the lowest abundance in plasma and binds strongly to mast cells, dominating the skin and submucosal layer. Elevated IgE levels are associated with parasitic infections such as worm and protozoan infections. Isotypes and subclasses can also reflect the progression and stage of infection, as seen in measles and human herpes virus 6 (HHV-6) changes in antibody populations during early and late infections.
Immune antibody libraries can be constructed in either a non-combinatorial or combinatorial manner. Non-combinatorial libraries use the original VH/VL pairing in single B cells, while combinatorial libraries rely heavily on a random combinatorial mix of heavy and light chain libraries. The antibody gene repertoire available in the immune environment provides a mixture of sequences present pre- and post-infection, with higher post-infection gene complexity due to affinity maturation. The repertoire from a post-infection environment will result in a repertoire that is biased against infection.
However, a combinatorial strategy increases the complexity of enriched gene representations by randomly pairing heavy and light chain proteins to produce the final immune library repertoire. This approach generates natural and unnatural VH/VL combinations which may result in functional or non-functional folding of clones within the library. In the context of antibody discovery, gene pairings with both natural and unnatural combinations of VH/VL may extend the functional diversity of the library. Alternatively, unnatural VH/VL pairings and CDR stacking could lead to misfolding, which could result in antibody aggregation and render them ineffective.
Mouse and human antibodies consist of two variable domains, and the same combinatorial diversity is not present in the immune libraries of camels and sharks, because antibodies adopt the single variable domain antibody (sdAb) format, known as VHH for camels and VNAR for sharks. These sdAbs, also known as nanoantibodies, contain only the VH domain and show some advantages in terms of solubility, stability, and target accessibility, mainly due to their small size.
However, in contrast to humans, the absence of the VL structural domain limits repertoire diversification, as the combinatorial pairing of VH and VL is not possible. Instead, the single variable domain immune library repertoire relies exclusively on germline diversification mechanisms that occur during immunization. The available repertoire consists of a combination of hyperimmune repertoire and natural repertoires available in immunized animals. Although smaller than mammalian diversity, the available repertoire is sufficient to generate high-affinity antibodies. The single-domain nature of sdAbs also means that only relatively small libraries are sufficient to represent immune complexes and simplify library generation.
Antibodies derived from immune antibody libraries have immense potential for diagnosis and therapy. Immune libraries created from recovered patients are ideal for therapeutic mAb, but immune libraries can also be created from immunized animals (such as mice, chickens, llamas, alpacas, camels, sheep, sharks, and non-human primates) for other applications. In bacterial infections, mAb is considered an alternative to antibiotics. The tetanus toxoid immune library, one of the first combinatorial immune antibody libraries constructed from the human peripheral B cell library, has successfully isolated several mAbs, but their full potential is still unknown. Subsequently, libraries of antibodies against botulinum toxin and immune antibodies against Pseudomonas aeruginosa have been developed. In the field of viral infections, the antibody library is also widely used to discover neutralizing monoclonal antibodies against dengue fever, Ebola virus disease, hepatitis B, human immunodeficiency virus infection, influenza, measles, and rabies. For parasitic infections, immune libraries can be used to treat parasite infections such as Plasmodium falciparum, taenia suis, Toxoplasma gondii, and Bacillus anthracis.
In conclusion, immune antibody library is not limited to producing only high-affinity disease-specific mAbs, but can also produce mAbs against other target proteins. As such, they are indispensable alternatives to naive libraries in antibody phage display development laboratories.
Reference
All listed services and products are For Research Use Only. Do Not use in any diagnostic or therapeutic applications.